- Node
- Similarity measures for ligand-binding conformations
Similarity measures for ligand-binding conformations
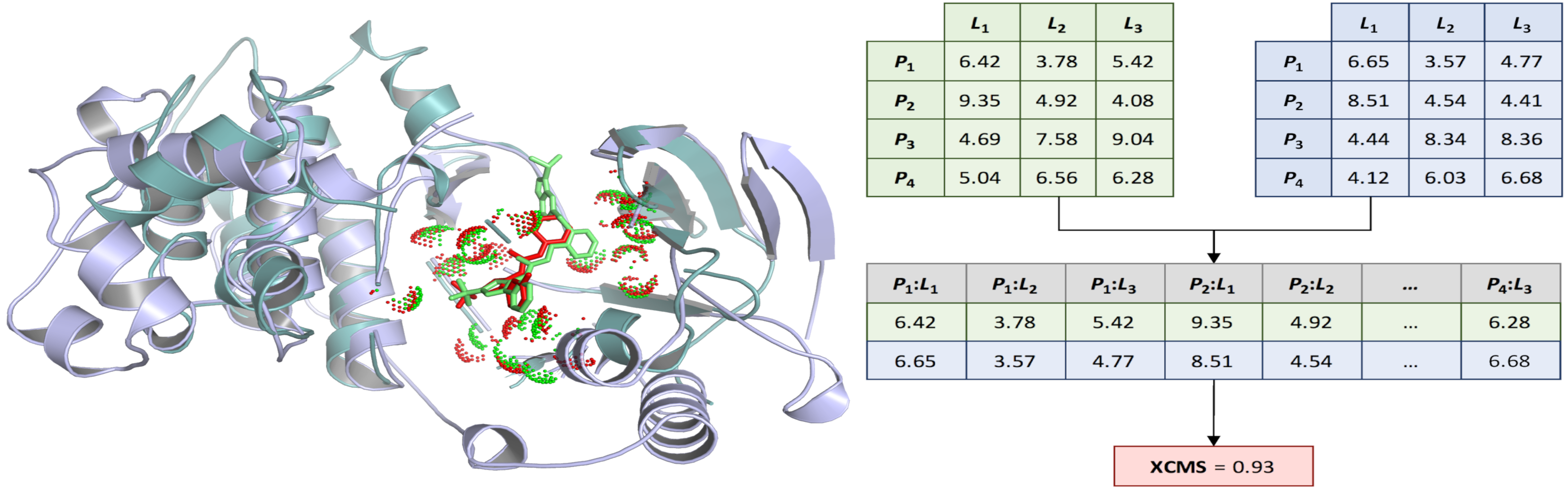
Structural and computational biologists often need to measure the similarity of ligand binding conformations. The commonly used root-mean-square deviation (RMSD) is not only ligand-size dependent, but also may fail to capture biologically meaningful binding features. To address these issues, we developed the Contact Mode Score (CMS), a new metric to assess the conformational similarity based on intermolecular protein-ligand contacts. The CMS is less dependent on the ligand size and has the ability to include flexible receptors. In order to effectively compare binding poses of non-identical ligands bound to different proteins, we further developed the eXtended Contact Mode Score (XCMS). CMS and XCMS provide a meaningful assessment of the similarity of ligand binding conformations.
