Ligand-binding site prediction
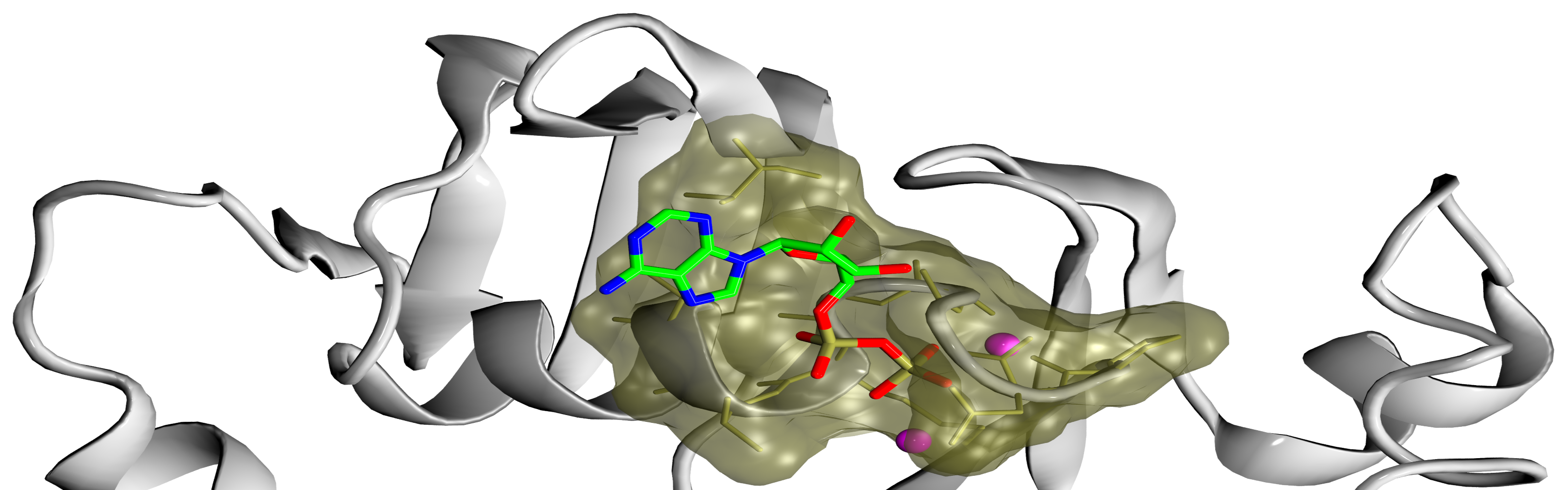
Much needed functional annotation of gene products across different species often requires the knowledge of protein-ligand interactions. Towards this goal, we developed eFindSite, an algorithm to efficiently identify ligand-binding sites and residues from weakly homologous templates with highly sensitive meta-threading, improved clustering techniques, and advanced machine learning methods. eFindSite can also be used to conduct ligand-based virtual screening employing consensus molecular fingerprints. Carefully calibrated confidence estimates strongly indicate that highly reliable ligand binding predictions are made for the majority of gene products in a given proteome, thus eFindSite holds a significant promise for large-scale genome annotation and drug development projects.
