High-performance computing
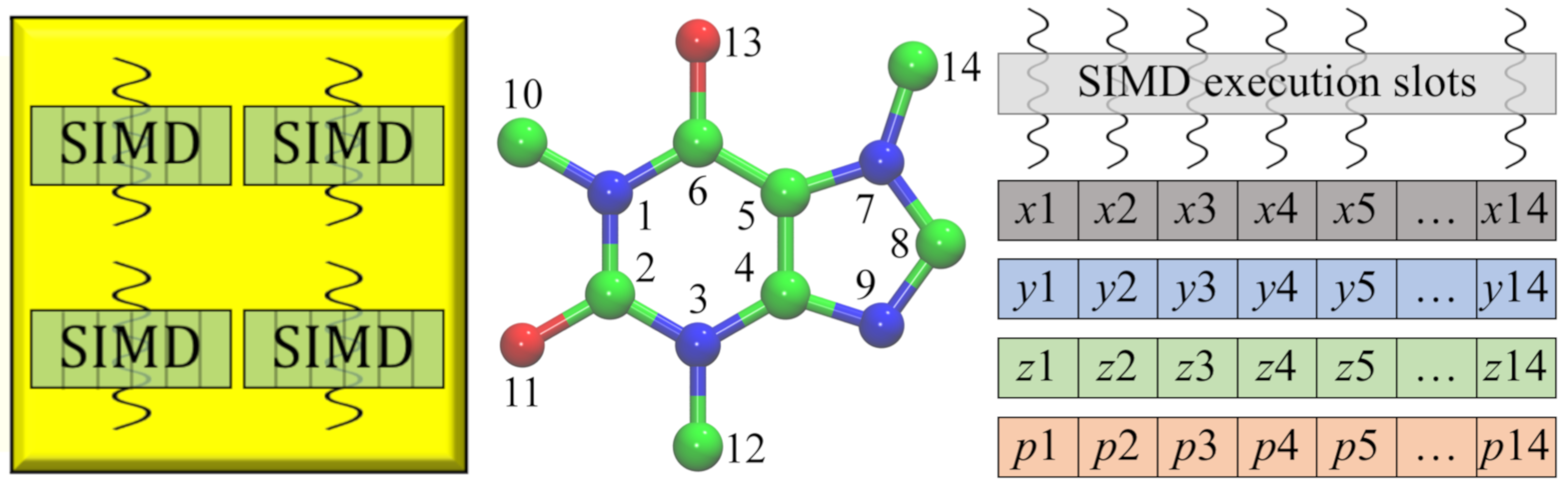
Drug development is routinely streamlined using computational approaches to improve hit identification and lead selection, enhance bioavailability, and reduce toxicity. New challenges arose because processing a large volume of data demands unprecedented computing resources. As part of a collaborative effort within LA-SiGMA, we ported several of our codes to heterogeneous computing platforms. For instance, GeauxDock can be deployed on multi-core Central Processing Units (CPUs) as well as massively parallel accelerators, Intel Xeon Phi and NVIDIA Graphics Processing Unit (GPU). Further, a parallel version of eFindSite was implemented mainly for the Intel Xeon Phi platform. These parallel codes yield significant performance improvements considerably accelerating their large-scale applications.
